Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Last updated 22 dezembro 2024

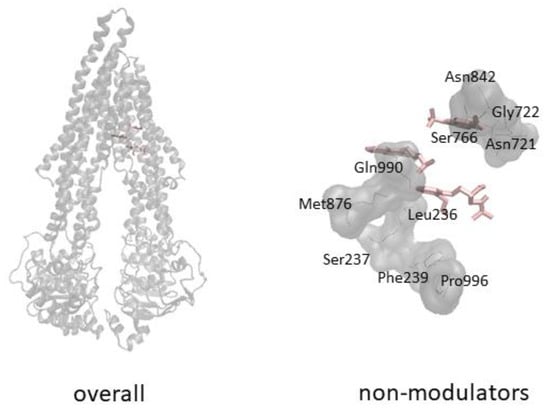
Cells, Free Full-Text

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

PDF] Computational models for predicting substrates or inhibitors of P- glycoprotein.

Effect of the Force Field on Molecular Dynamics Simulations of the Multidrug Efflux Protein P-Glycoprotein

Computational Biology and Chemistry in MTi: Emphasis on the Prediction of Some ADMET Properties - Miteva - 2017 - Molecular Informatics - Wiley Online Library
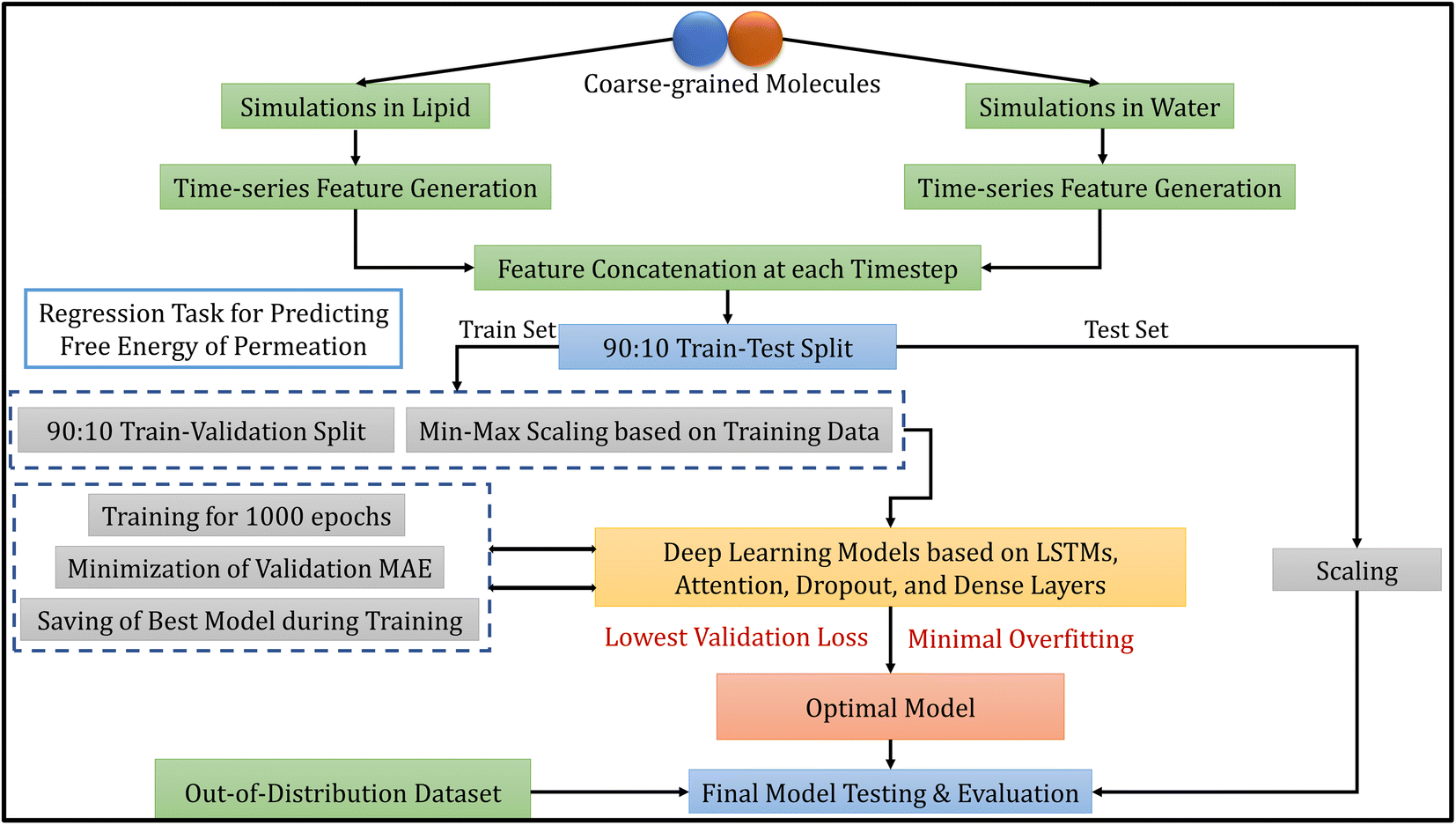
Deep learning models for the estimation of free energy of permeation of small molecules across lipid membranes - Digital Discovery (RSC Publishing) DOI:10.1039/D2DD00119E

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

IJMS, Free Full-Text

Computational and artificial intelligence-based approaches for drug metabolism and transport prediction: Trends in Pharmacological Sciences
Recomendado para você
-
 Brain Test Level 372 Walkthrough22 dezembro 2024
Brain Test Level 372 Walkthrough22 dezembro 2024 -
 Brain Sciences, Free Full-Text22 dezembro 2024
Brain Sciences, Free Full-Text22 dezembro 2024 -
 DOP 2 Level 372 Answer the phone Answer - Daze Puzzle22 dezembro 2024
DOP 2 Level 372 Answer the phone Answer - Daze Puzzle22 dezembro 2024 -
 Relative Selectivity of Covalent Inhibitors Requires Assessment of Inactivation Kinetics and Cellular Occupancy: A Case Study of Ibrutinib and Acalabrutinib22 dezembro 2024
Relative Selectivity of Covalent Inhibitors Requires Assessment of Inactivation Kinetics and Cellular Occupancy: A Case Study of Ibrutinib and Acalabrutinib22 dezembro 2024 -
 Microbiota‐gut‐brain axis as a regulator of reward processes - García‐Cabrerizo - 2021 - Journal of Neurochemistry - Wiley Online Library22 dezembro 2024
Microbiota‐gut‐brain axis as a regulator of reward processes - García‐Cabrerizo - 2021 - Journal of Neurochemistry - Wiley Online Library22 dezembro 2024 -
 Он хочет быть выше. 372 уровень Brain Test22 dezembro 2024
Он хочет быть выше. 372 уровень Brain Test22 dezembro 2024 -
 Brain test level 372 He wants big muscles22 dezembro 2024
Brain test level 372 He wants big muscles22 dezembro 2024 -
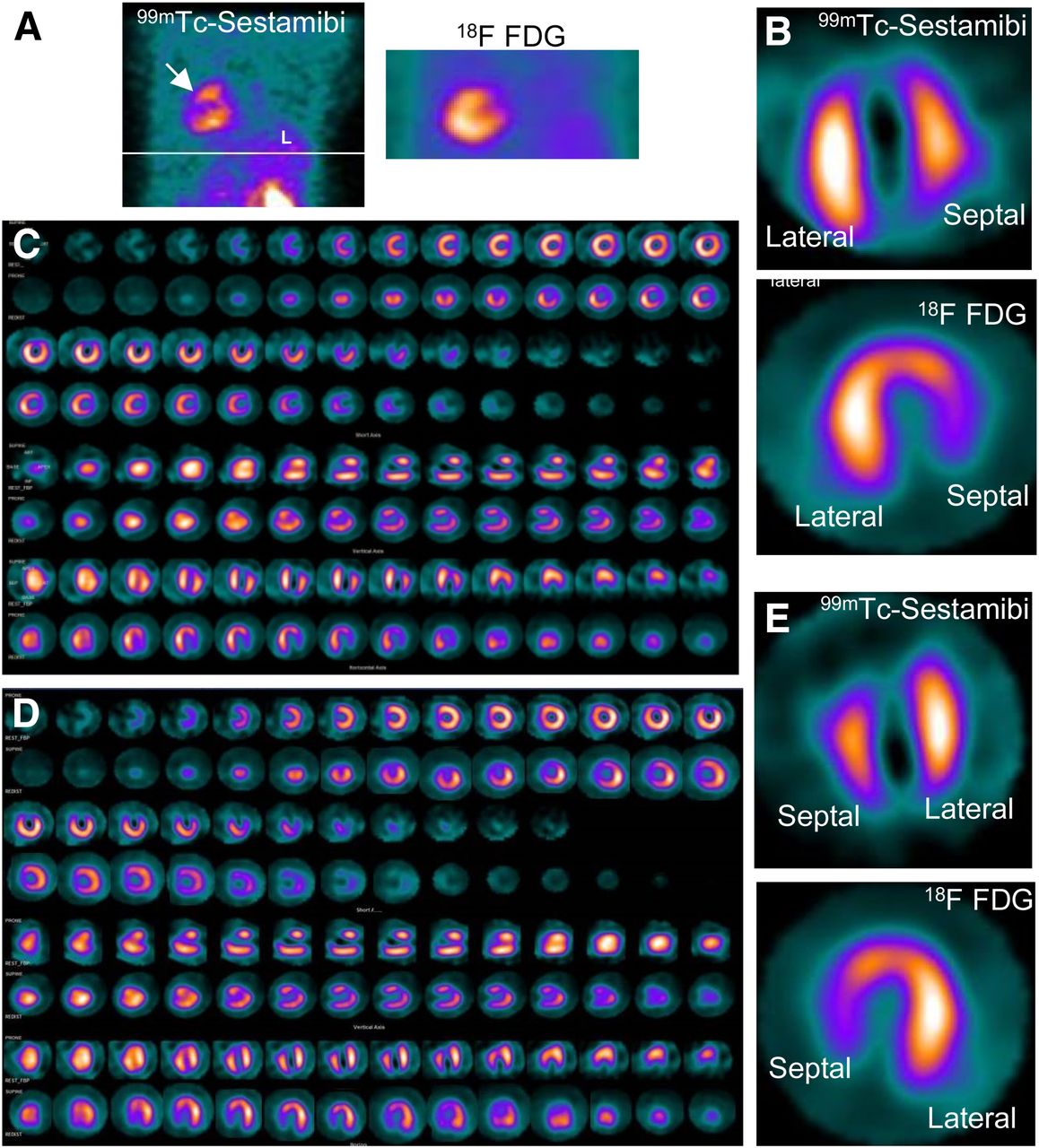 Assessment of Myocardial Viability Using Nuclear Medicine Imaging in Dextrocardia22 dezembro 2024
Assessment of Myocardial Viability Using Nuclear Medicine Imaging in Dextrocardia22 dezembro 2024 -
SEVEN SEAS Multivitamin + Cod Liver Oil Syrup 100ml. *BENEFITS* - Improves Appetite - Improves Memory - Strengthens Bones & Teeth - Boosts the Immune, By Seven Seas Pharmacy22 dezembro 2024
-
 The answer to level 371, 372, 373, 374, 375, 376, 377, 378, 379 and 380 is DOP 2: Delete One Part - Brain Game Master22 dezembro 2024
The answer to level 371, 372, 373, 374, 375, 376, 377, 378, 379 and 380 is DOP 2: Delete One Part - Brain Game Master22 dezembro 2024
você pode gostar
-
 BRITISH SUMMER TIME BEGINS - March 31, 2024 - National Today22 dezembro 2024
BRITISH SUMMER TIME BEGINS - March 31, 2024 - National Today22 dezembro 2024 -
 G1 - Hackers conseguem rodar aplicativos iOS na televisão usando Apple TV - notícias em Tecnologia e Games22 dezembro 2024
G1 - Hackers conseguem rodar aplicativos iOS na televisão usando Apple TV - notícias em Tecnologia e Games22 dezembro 2024 -
 Subtitles be like : r/Boruto22 dezembro 2024
Subtitles be like : r/Boruto22 dezembro 2024 -
 Página 4 Fotos Perfil Anime, 83.000+ fotos de arquivo grátis de22 dezembro 2024
Página 4 Fotos Perfil Anime, 83.000+ fotos de arquivo grátis de22 dezembro 2024 -
 sanrio boys by viodiba on DeviantArt22 dezembro 2024
sanrio boys by viodiba on DeviantArt22 dezembro 2024 -
 Toppers, Labels or Stickers for Free Print.22 dezembro 2024
Toppers, Labels or Stickers for Free Print.22 dezembro 2024 -
 Love Heart Dona Tee Grunge/Vintage Style Black Dona22 dezembro 2024
Love Heart Dona Tee Grunge/Vintage Style Black Dona22 dezembro 2024 -
 New Sword Art Online FULLDIVE First Limited Edition 2 DVD Booklet Japan22 dezembro 2024
New Sword Art Online FULLDIVE First Limited Edition 2 DVD Booklet Japan22 dezembro 2024 -
ipop_la has been great. Been training for acting & modeling with workshops/bootcamps. Prepping for some big auditions 😎 #iPop has this…22 dezembro 2024
-
 Análise: Dragon Ball Z: Kakarot (Multi) traz a experiência mais imersiva de Goku e seus amigos - GameBlast22 dezembro 2024
Análise: Dragon Ball Z: Kakarot (Multi) traz a experiência mais imersiva de Goku e seus amigos - GameBlast22 dezembro 2024

